Preference point-based NSGA-II¶
Example¶
[1]:
from jmetal.algorithm.multiobjective.nsgaii import NSGAII
from jmetal.operator.crossover import SBXCrossover
from jmetal.operator.mutation import PolynomialMutation
from jmetal.problem import ZDT2
from jmetal.util.comparator import GDominanceComparator
from jmetal.util.termination_criterion import StoppingByEvaluations
problem = ZDT2()
max_evaluations = 25000
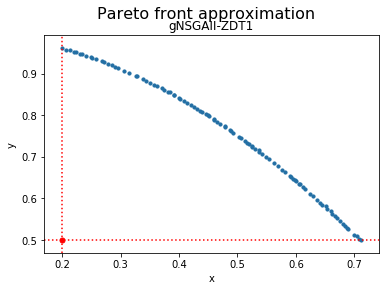
reference_point = [0.2, 0.5]
algorithm = NSGAII(
problem=problem,
population_size=100,
offspring_population_size=100,
mutation=PolynomialMutation(probability=1.0 / problem.number_of_variables, distribution_index=20),
crossover=SBXCrossover(probability=1.0, distribution_index=20),
dominance_comparator=GDominanceComparator(reference_point),
termination_criterion=StoppingByEvaluations(max=max_evaluations)
)
algorithm.run()
solutions = algorithm.get_result()
We can now visualize the Pareto front approximation:
[3]:
from jmetal.lab.visualization.plotting import Plot
from jmetal.util.solution import get_non_dominated_solutions
front = get_non_dominated_solutions(solutions)
plot_front = Plot(plot_title='Pareto front approximation', axis_labels=['x', 'y'], reference_point=reference_point)
plot_front.plot(front, label='gNSGAII-ZDT1')
API¶
- class jmetal.algorithm.multiobjective.nsgaii.DynamicNSGAII(problem: ~jmetal.core.problem.DynamicProblem[~jmetal.algorithm.multiobjective.nsgaii.S], population_size: int, offspring_population_size: int, mutation: ~jmetal.core.operator.Mutation, crossover: ~jmetal.core.operator.Crossover, selection: ~jmetal.core.operator.Selection = <jmetal.operator.selection.BinaryTournamentSelection object>, termination_criterion: ~jmetal.util.termination_criterion.TerminationCriterion = <jmetal.util.termination_criterion.StoppingByEvaluations object>, population_generator: ~typing.Generator = <jmetal.util.generator.RandomGenerator object>, population_evaluator: ~jmetal.util.evaluator.Evaluator = <jmetal.util.evaluator.SequentialEvaluator object>, dominance_comparator: ~jmetal.util.comparator.DominanceComparator = <jmetal.util.comparator.DominanceComparator object>)[source]¶
Bases:
NSGAII[S,R],DynamicAlgorithm
